Abstract
Introduction: Gemtuzumab ozogamicin (GO) has re-emerged as a potential agent with promise in acute myeloid leukemia (AML) treatment. GO received accelerated approval in 2000 for treatment of relapsed AML in older patients but was withdrawn from the market due to lack of benefit and increased early mortality in SWOG S0106 trial. Several studies thereafter demonstrated GO improved clinical outcomes with pronounced effects in patients with favorable-risk cytogenetics. In light of new results from several randomized trials, an FDA advisory panel recently determined that GO has a favorable risk/benefit profile as an adjunctive therapy for certain newly-diagnosed AML patients. We recently reported that CD33 splicing polymorphism rs12459419 C>T regulates production of alternatively spliced CD33 isoform lacking exon 2 and hence the IgV domain for CD33(Lamba et al, 2017). Consistent with this, we demonstrated that rs12459419 was significant predictor of cell surface CD33 intensities as well as clinical response in the randomized COG-AAML0531 trial. We now expand our investigation to other potentially significant CD33 SNPs for impact on GO efficacy in de novo AML patients enrolled in the same AAML0531 trial.
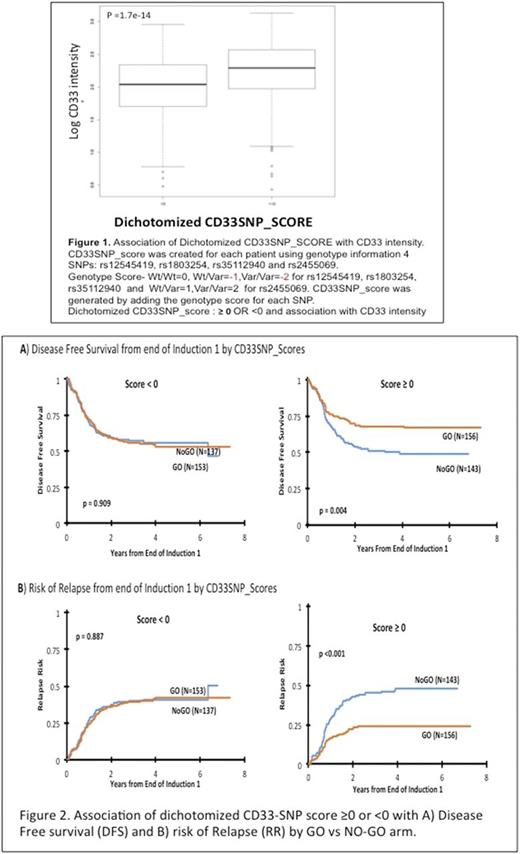
Methods: We genotyped five CD33 SNPs: rs2455069 (Arg60Gly), rs35112940 (Arg304Gly), rs61736475 (Ser305Pro) and rs1803254 and associated these with CD33 cell surface levels and clinical outcome in AML patients (n=817: 408=No-GO and 409=GO). We further developed a composite CD33 SNP-score ranging from -6 to +2, using genotype information on the 4-CD33 SNPs with significant association with cell surface CD33 levels. A directional genotype score was generated for each SNP by taking into account the direction of association of variant allele with CD33 levels. Genotype score ranged from 0 to -2 for rs12459419, rs1803254 and rs35112940 -with variant alleles associated with lower CD33 intensity. For CD33 SNP rs2455069 -with variant allele associated with high CD33 cell surface intensity, genotype scores ranged from 0 to +2 based on number of variant alleles present. Composite CD33-SNP Score = Σ (individual genotype scores of rs12459419, rs1803254, rs2455069 and rs35112940) followed by grouping into score of ≥0 OR <0.
Results: CD33 coding SNPs-rs2455069, rs35112940 and 3'UTR SNP-rs1803254 but not rs61736475 were associated with cell surface CD33 levels on AML blasts and superior response with GO. These SNPs did not show any linkage with each other and the effect seen was independent. The CD33-SNPscore was therefore built using genotype scores for previously reported rs12459419 and the new rs2455069, rs1803254 and rs35112940 CD33 SNPs. Patients with a CD33-SNP score of ≥0 had higher cell surface CD33 levels (Figure 1) and demonstrated improved 5-year disease-free survival of 67±8% vs. 49±9% (hazard ratio [HR]=0.58, [95% confidence interval 0.41-0.84]; p=0.004) as well as reduced risk of relapse (24±7% vs. 48±9%; HR=0.448 [95% CI: 0.30-0.67]; p <0.001, Figure 2) when treated with GO vs. NO-GO. GO did not improve outcomes in patients with a CD33-SNPscore of <0, which was associated with lower CD33 expression levels. Subgroup analyses showed similar results, in low- and intermediate-risk patients, but for high-risk group statistical significance was not reached.
Conclusion: In this study to comprehensively evaluate the role of CD33 SNPs on outcome we created a composite CD33-SNP_score using a directional genotype score for these three and our previously reported splicing SNP, rs12459419. Our findings suggest the use of GO with intensive induction chemotherapy in childhood AML should be restricted to patients with CD33-SNP score of ≥0. Although CC genotype for our recently reported splicing SNP rs12459419 was significantly represented in CD33-SNP score of ≥ 0, it is important to realize it occurs with an allele frequency of ~0.3 in European patients but is less frequent in other ethnic groups (ranging from ~0.10-0.15 in African American and Asian and non-existent in Sub-Saharan Africans), whereas rs1803254 is more frequent in black vs. white patient (0.32 vs. 0.15), thus the use of a composite CD33-SNP score would broaden identification of those patients who will benefit from GO or other CD33 targeted agents, in non-European populations. Validation of this CD33-SNP score with GO response remains to be performed and if reproduced, future studies should consider the use of the CD33-SNP score to guide GO therapy.
Loken: Hematologics Inc: Employment, Equity Ownership. Walter: ADC Therapeutics: Research Funding; Aptevo Therapeutics: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal